Within the PrevOP research network "Preventing the progression of primary Osteoarthritis by high impact long-term Physical exercise regimen – key mechanisms, efficacy, and long-term results." the aim of sub-project 4 is to assess and to quantitatively analyze morphological and structural changes related to osteoarthritis (OA) with respect to different levels of exercise. In arthritic knees typical OA-related findings are the degeneration of cartilage, menisci, and ligaments. Furthermore, in the majority of cases of severe OA, bone marrow lesions and subarticular cysts occur. It is assumed that these structures can be quantitatively assessed with medical imaging techniques, in particular MRI (Fig. 1). In this project, using experts' reading and machine learning techniques, OA-related quantitative image features will be identified for every relevant structure. Combining all image features might lead to a novel image-based OA-score. This OA-score could be used to support the hypothesis that exercise slows down the progression of OA.
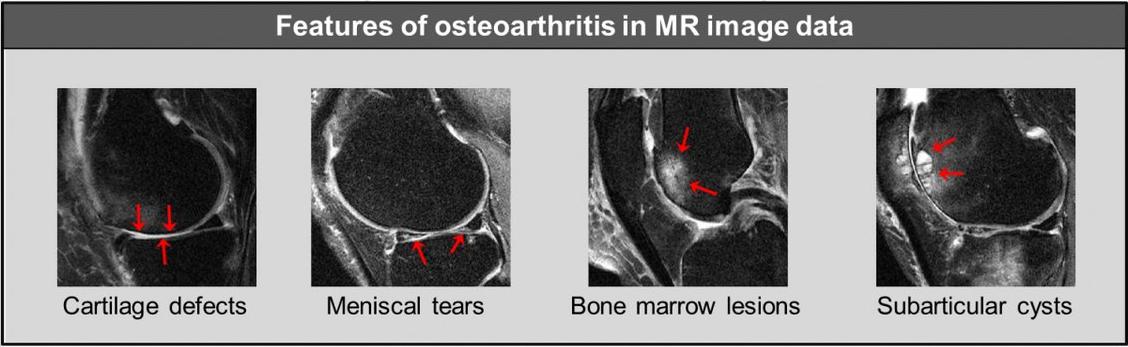
Fig 1: Features of osteoarthritis: Cartilage defects, meniscal tears, bone marrow lesions and subarticular cysts.
Identifying OA-related features with data of the Osteoarthritis Initiative (OAI) database
In the OAI database MRIs and radiographs are publicly available for almost 5.000 patients. In addition to basic scoring data (i.e. WOMAC and Kellgren-Lawrence score) several image assessment studies have been carried out for OAI data:
- Whole-Organ Magnetic Resonance Imaging Score (WORMS) and Boston Leeds Osteoarthritis Knee Score (BLOKS) scoring results have been published in Mai 2011 for 115 patients
- MRI Osteoarthritis Knee Score (MOAKS) reading results have been published in April 2015 for 600 patients
In this project, image features will be extracted for each OA-related structure (cartilage, menisci, ligaments, bone marrow lesions and subarticular cysts). Employing methods of machine learning and using the data of the WORMS, BLOKS and MOAKS image assessment studies, the correlation between these image features and the experts' reading will be investigated. Features, which were identified as being essential for OA progression, will be combined to an image-based OA score (Fig. 2).
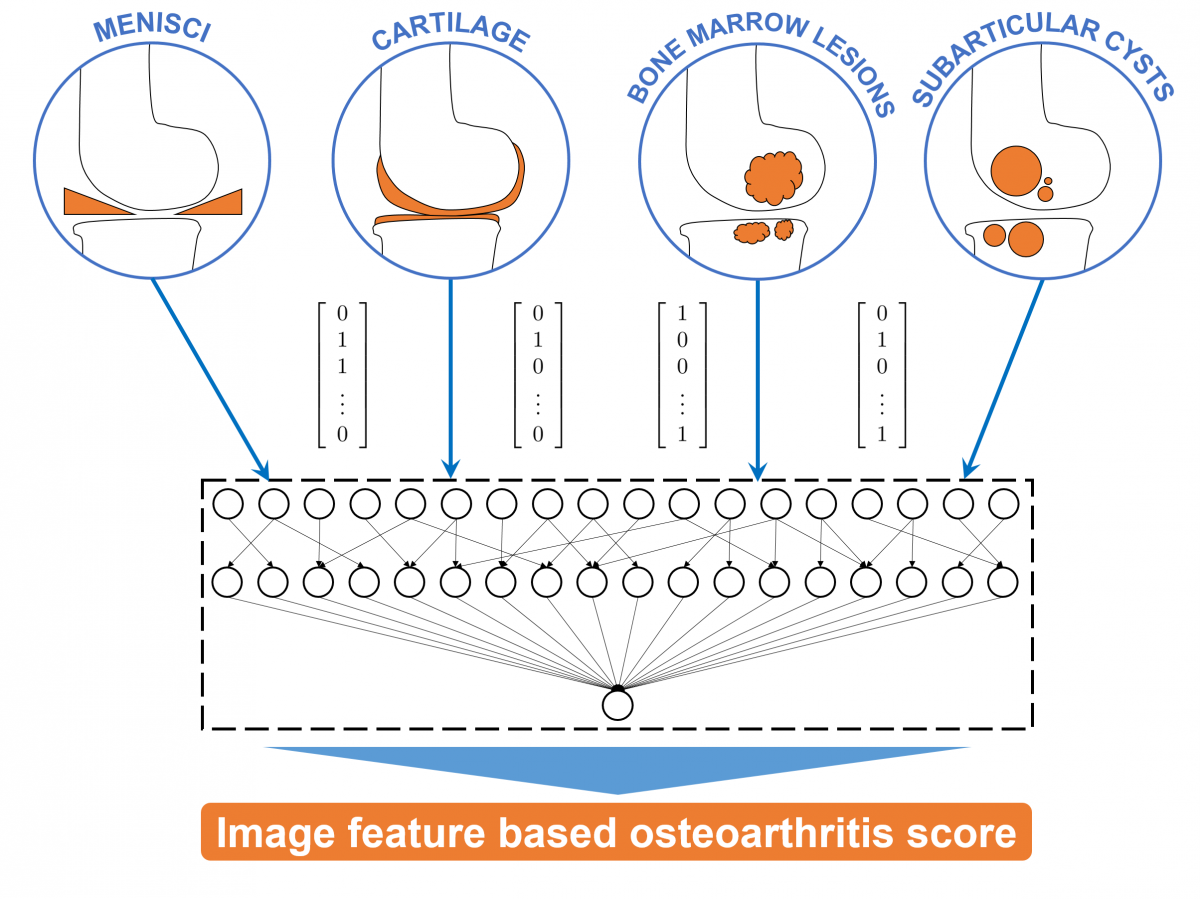
Fig 2: Image-based quantitative features are extracted for the cartilage, menisci, bone marrow lesions and subarticular cysts. These image features are combined to a novel osteoarthritis score.
Automated segmentation of knee bones, cartilage, and menisci
To generate specific regions of interest (ROI) for the considered anatomical structures in the MRI data, the knee bones (femur and tibia), the femoral and tibial cartilage [1], and the menisci [2] are automatically segmented using methods based on convolutional neural networks and statistical shape models.
Analysis of the knee menisci
Using the 3D segmentations of the menisci and the knee bones quantitative measurements (meniscal volume, meniscal extrusion, and tibial coverage) were calculated [2]. We investigated the correlation of these measurements and the OA grade using data from the OAI database [2].
Also, we pursued the aim to detect meniscal tears in MRI data by using deep learning methods with the automatically determined meniscal ROI and MOAKS semi-quantitative meniscal morphology reading [3].
Analysis of the knee bones and cartilage
An deep learning based method for the automated segmentation of knee bones and cartilage was developed and thoroughly validated [1]. We will investigate the usability of cartilage volume and thickness measures derived from these segmentations as suitable and reliable OA biomarkers. We employed a multi-loss function to further increase the accuracy of cartilage volume measurements [4].
Analysis of the knee alignment
A deep learning based method for the automated assessment of knee alignment from full-leg X-Ray images was developed [5]. The accuracy was evaluated for over 3,000 datasets from the OAI database.
Data availability
All generated data are available at pubdata.zib.de.
References
[1] F Ambellan, A Tack, M Ehlke, S Zachow (2019). Automated Segmentation of Knee Bone and Cartilage combining Statistical Shape Knowledge and Convolutional Neural Networks: Data from the Osteoarthritis Initiative. Medical Image Analysis; 52(2), p. 109--118
[2] A Tack, A Mukhopadhyay, S Zachow (2018). Knee Menisci Segmentation using Convolutional Neural Networks: Data from the Osteoarthritis Initiative. Osteoarthritis and Cartilage; 26(5), p. 680--688.
[3] V Reddy, A Tack, B Preim, S Zachow (2017). Automatic Classification of 3D MRI data using Deep Convolutional Neural Networks. Masters thesis, Otto-von-Guericke-Universität Magdeburg
[4] A Tack, S Zachow (2019). Accurate Automated Volumetry of Cartilage of the Knee using Convolutional Neural Networks: Data from the Osteoarthritis Initiative. ISBI 2019
[5] A Tack, B Preim, S Zachow (2021). Fully automated Assessment of Knee Alignment from Full-Leg X-Rays employing a "YOLOv4 And Resnet Landmark regression Algorithm" (YARLA): Data from the Osteoarthritis Initiative. Computer Methods and Programs in Biomedicine; epub ahead of print